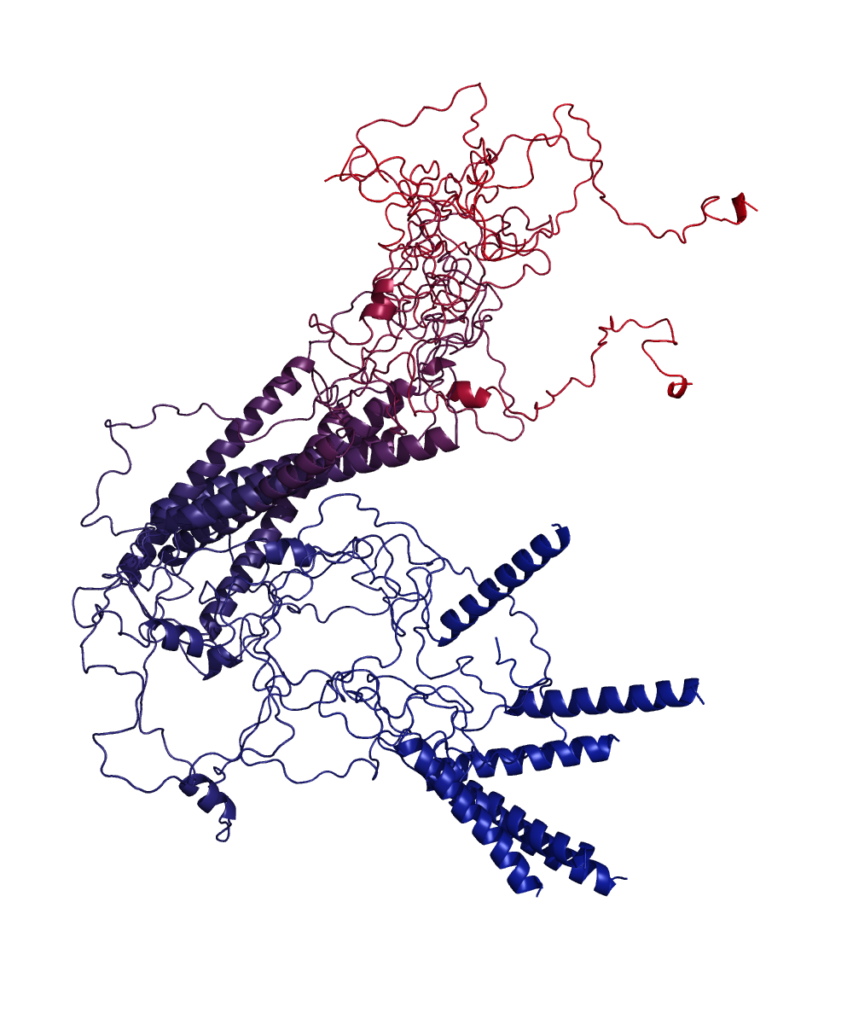
Non-globular proteins in the era of Machine Learning
COST Action CA21160
THE ACTION
ABOUT US
The ML4NGP COST Action aims to establish an interdisciplinary pan-European network to address non-globular proteins, a fundamental research gap in the protein science field. this COST Action aims to describe the function and properties of Non-globular Proteins (NGPs) by combining experimental data and novel machine learning (ML) approaches.
Scientific Background
Protein structure prediction has long been considered the “Holy Grail” of structural biology. The recent success of AI-based tools (e.g. AlphaFold) has ushered in a new era of highly accurate structure prediction, bringing to light the secrets hidden in the three-dimensional structures of globular proteins, increasing our understanding about their structural features and molecular function. However, a large proportion of the proteomes from all domains of life are rich in sequences that do not fold into regular structures, commonly known as non-globular proteins (NGPs). NGPs comprise intrinsically disordered regions, repeats, low-complexity sequences, aggregation-prone and phase-separating sequences, and are implicated in a range of age-related diseases. These proteins have shaken the long-held structure-function paradigm where well-defined native protein structures are needed for function.
Their heterogeneous structural states and low sequence complexity challenge current experimental structure determination techniques and machine learning (ML) methods for structure prediction, making the molecular understanding of their sequence-structure-dynamics-function relationship difficult. The recent improvements of ML approaches and advances in determining NGP structural ensembles call for a timely re-assessment of the interplay between experiments and computation.



OUR VISION
The Action aims to strengthen both sides, in a way that machine learning can be used to enhance the combination of experimental methods and computational approaches to study NGPs. This Action aims that experimental frameworks are designed to provide information to computational methods, and computational methods are developed, trained and benchmarked with experimental data.
This integrative approach will provide new frameworks and methodologies to understand and to study and quantify the relationship between sequence-structure-dynamics-function of NGPs, stimulated by the development of new computational methods, standardization of best-practices for NGP experimental and computational research and the generation of novel curated datasets.
Action Details
CSO Approval date
27/05/2022
Start date
25/10/2022
End date
24/10/2026
How to participate in the Action?
Inform the Action Chair of your interest (email).
Apply to join your Working Groups of interest.
Management Committee nominations are carried out through the COST National Contact Point.
Latest News
-
 02 Jun 2025Advancing the Science of Non-Globular Proteins: The 3rd ML4NGP Meeting in Vilnius
02 Jun 2025Advancing the Science of Non-Globular Proteins: The 3rd ML4NGP Meeting in Vilnius -
 12 Feb 20253rd ML4NGP Training School in Brno: A Week of Intensive Learning and Collaboration
12 Feb 20253rd ML4NGP Training School in Brno: A Week of Intensive Learning and Collaboration -
 22 Oct 2024Alexander Monzon Represents ML4NGP COST Action at AccelNet 6th Annual Meeting in Washington DC
22 Oct 2024Alexander Monzon Represents ML4NGP COST Action at AccelNet 6th Annual Meeting in Washington DC -
 04 Jun 2024Successful Conclusion of the ML4NGP Meeting in Thessaloniki: A Hub of Cutting-Edge Research and Collaboration
04 Jun 2024Successful Conclusion of the ML4NGP Meeting in Thessaloniki: A Hub of Cutting-Edge Research and Collaboration -
 07 Mar 20242nd ML4NGP Training School Achieves Another Remarkable Success
07 Mar 20242nd ML4NGP Training School Achieves Another Remarkable Success