Three weeks ago, the vibrant city of Thessaloniki, Greece, served as the backdrop for the highly anticipated second meeting on Machine Learning for Non-Globular Proteins, co-organized by ML4NGP COST Action and MSCA-RISE REFRACT consortia. This landmark event brought together nearly 130 participants from 34 countries, fostering a global exchange of knowledge and innovation in the field of machine learning and protein science.
The conference was graced by four distinguished keynote speakers who set the tone with their groundbreaking research and visionary insights.
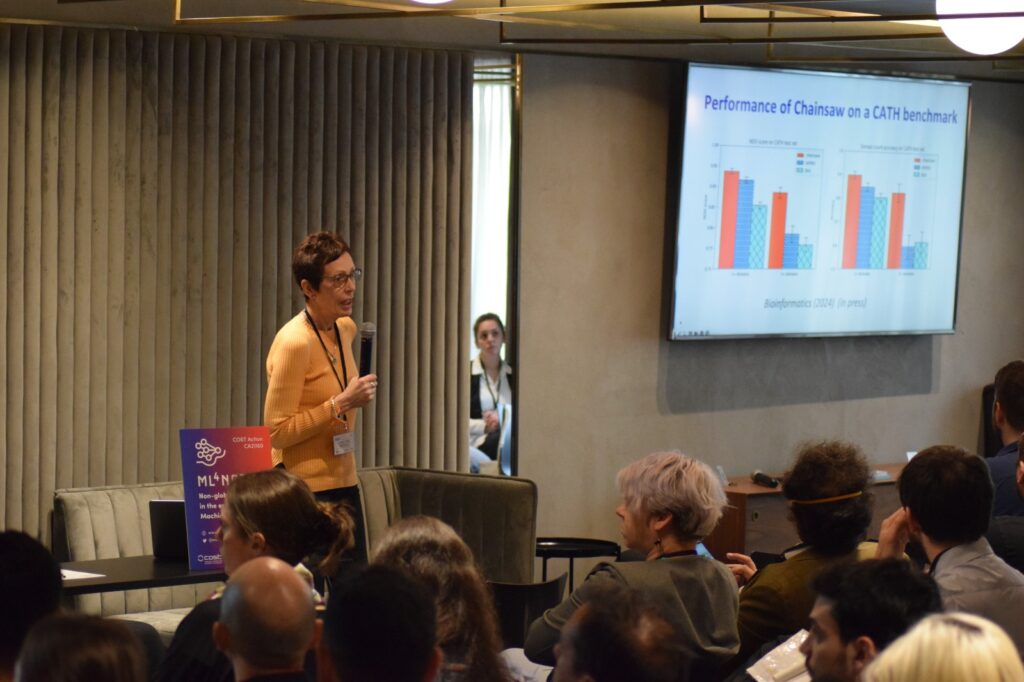
Christine Orengo introduced powerful new machine learning algorithms designed to capture intricate relationships between protein structures, sequences, and functions. Her talk emphasized the transformative potential of these algorithms in understanding complex biological systems.

Michele Vendruscolo tackled the formidable challenges of drug discovery for intrinsically disordered proteins (IDPs) associated with amyloid-related diseases. He provided a hopeful outlook on the therapeutic possibilities, shedding light on innovative approaches to target these elusive proteins.
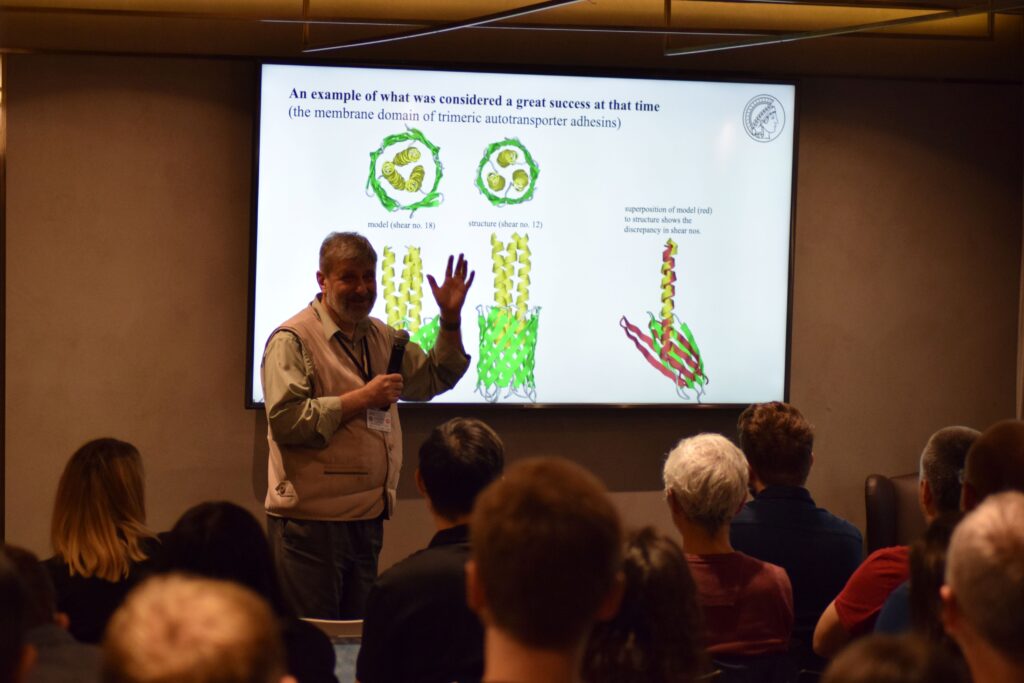
Andrei Lupas delivered an engaging and inspiring talk on the Critical Assessment of Structure Prediction (CASP) and the concept of the virtual proteome of life, underscoring the importance of accurate protein structure prediction in modern biology.

Christos Ouzounis provided a comprehensive overview of the evolution of bioinformatics, from studying globular proteins to understanding the structure-function relationships of IDPs over the past two decades. His talk highlighted the significant strides made in the field and the promising future ahead.
The conference featured a top-level scientific program, with more 5 selected talks and 28 short talks. Additionally, 64 posters were presented, with 11 chosen for flash poster presentations. The diverse range of topics covered the interdisciplinary and innovative research in the field of machine learning for non-globular proteins.





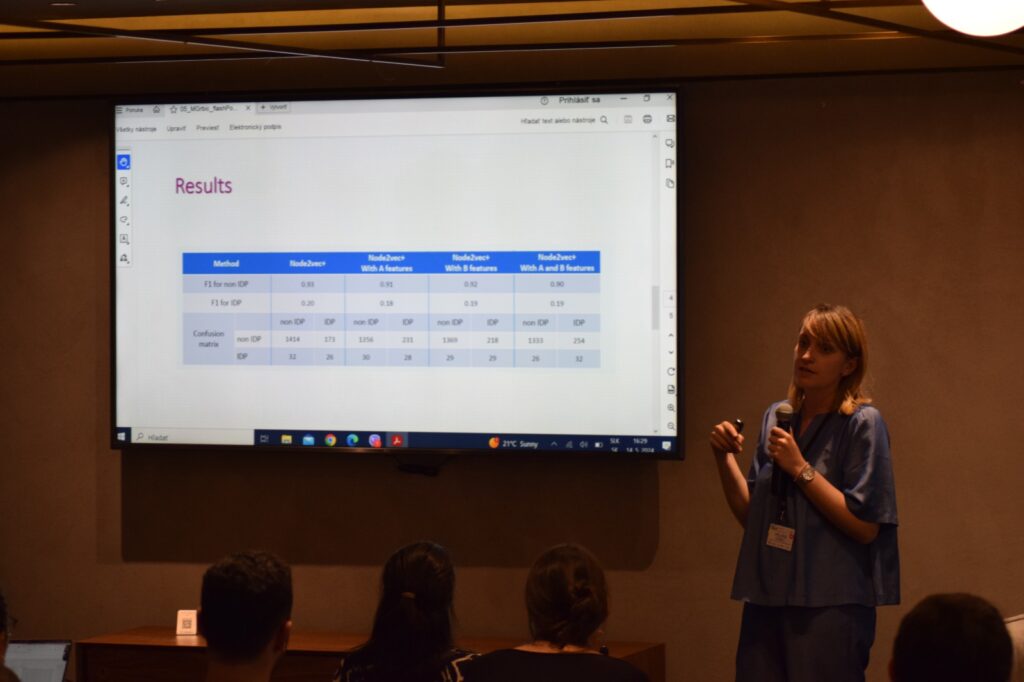

Photos from speakers at the 2nd ML4NGP Meeting at ONOMA Hotel, in Thessaloniki – Greece.
The meeting set the stage to a global exchange of knowledge covering a wide range of topics, reflecting the dynamic and multifaceted nature of protein research:
- A novel repository of anti-aSyn peptides was described, aimed at identifying therapeutic agents or biomarkers for Parkinson’s disease.
- The power of NMR in studying the mechanisms of spider silk fiber formation was demonstrated, revealing its application in studying protein dynamics and mechanisms.
- Innovative machine learning frameworks were presented for improved prediction of protein interaction sites, leveraging the capabilities of AlphaFold2-Multimer.
- Resources were introduced that provide processed, harmonized, and integrated bioactivity data on small molecules, enhancing the accessibility and usability of this critical information.
- Deep learning-based Markov state modeling approaches were discussed, providing insights into the characterization of folding-upon-binding pathways in proteins.
- An inspiring talk on the Gaussian Network Model (GNM)-based Transfer Entropy (TE) approach revealed multi-directional allosteric pathways within protein structures.
The conference also highlighted significant advancements in the development of computational tools and models. For instance, a new latent diffusion model for constructing IDR ensembles was introduced, and a coarse-grained model of IDRs, named CALVADOS, was presented to create a proteome-wide database of conformational ensembles. These tools are poised to enhance our understanding of sequence-ensemble relationships and the biological roles of IDPs.
The scientific committee recognized the outstanding contributions of young researchers with four awards for the best posters. These awards celebrated innovative research and excellence of PhD students, highlighting their contribution with cutting-edge work in the field of machine learning for non-globular proteins.



Photos of poster sessions and the four awardees with the best poster presentation.
During the conference, dedicated working group (WG) meetings were held to engage participants actively in shaping the future directions of the ML4NGP COST Action. These discussions facilitated collaboration and the exchange of ideas, pushing forward the next steps for each working group.
By integrating these discussions into the conference program, the meeting successfully aligned the goals of the working groups with the broader objectives of the ML4NGP Action, setting a clear path for future research and collaboration and for science dissemination. Ultimately, the collective efforts from these meetings highlighted the importance of fostering collaborations between computational and experimental researchers in this field. The main take home messages were the need for improvement of experimental data annotation and of communication between experimental and computational researchers, through setting an array of guidelines and common vocabulary to further advance the collaborative research between investigators of various backgrounds.




Participants engaged into insightful and dynamic discussions during the talks and the WG meetings.
The success of the ML4NGP meeting would not have been possible without the dedication and hard work of the local organizing team, led by Fotis Psomopoulos. The hospitality of the ONOMA Hotel in Thessaloniki also played a crucial role in providing a conducive environment for fruitful discussions and networking.
In conclusion, the ML4NGP meeting in Thessaloniki was a resounding success, bringing together a diverse group of experimental and computational researchers to share their latest findings and forge new collaborations. The insights and innovations presented at the conference underscore the pivotal role of machine learning in advancing our understanding of non-globular proteins, paving the way for future discoveries and therapeutic breakthroughs.

Group photo with the participants and speakers of the second ML4NGP meeting held in Thessaloniki, Greece from 14 to 17 May 2024.
You can find more information about our meeting on the 2nd edition of ML4NGP Newsletter.
What people are saying about the meeting:

I really enjoyed the keynote presentations, particularly because they provided deep insights into the latest advancements and applications of machine learning for non-globular proteins.
Anonymous

The ML4NGP 2024 meeting was an outstanding introduction to the world of protein disorder. I have rarely met such a passionate and focused academic community, and the impact and urgency of understanding protein disorder was made very clear through the compelling talks and student posters. I left the conference with a new perspective on how machine learning might play a role in this topic, and optimism that this strong community is poised to make breakthroughs in this space.
Joshua Pan Google DeepMind