Non-globular proteins in the era of Machine Learning
COST Action CA21160
CONFERENCE
THIS EVENT HAS PASSED.
1st ML4NGP MEETING on Machine Learning and Non-globular proteins
July 5-7, 2023
Bratislava, Slovakia



welcome message
Non-globular proteins (NGPs) heterogeneous structural states and low sequence complexity challenge current experimental structure determination techniques and machine learning (ML) methods for structure prediction, making the molecular understanding of their sequence-structure-dynamics-function relationship difficult. The recent improvements of ML approaches and advances in determining NGP structural ensembles call for a timely re-assessment of the interplay between experiments and computation.
The 1st ML4NGP MEETING 2023 will be held from July 5 to 7 in Bratislava providing an opportunity for computational researchers and experimentalists interested in adapting Machine Learning tools and advancements for the study of NGPs to present and discuss current developments in the field.
This meeting is organized in the framework of COST ACTION CA21160 Non-globular proteins in the era of Machine Learning – ML4NGP, IDPFun (“Driving the functional characterization of intrinsically disordered proteins”) and REFRACT (“Repeat protein Function, Refinement, Annotation and Classification of Topologies”). It is hosted by the Institute of Neuroimmunology, Slovak Academy of Sciences.
The program includes a pre-event meeting on July 4, which is exclusive for ML4NGP Core Group members.

|
Deadline |
Status |
|
|
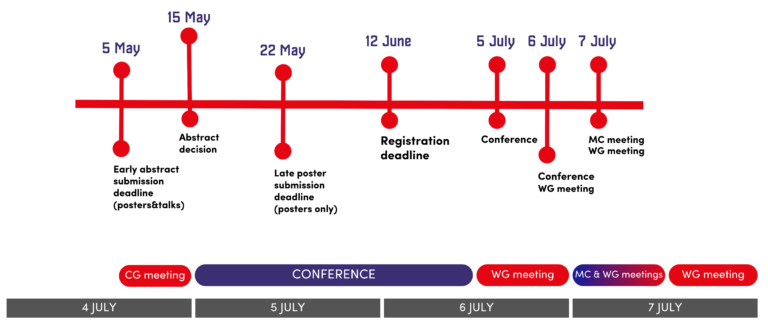
Abstract submission deadline (posters & talks) |
May 5, 2023 |
Closed |
|
Abstract decision |
May 15, 2023 |
Closed |
|
Late abstract submission deadline (posters only) |
May 22, 2023 |
Closed |
|
Early registration deadline |
June 12, 2023 |
Closed |
|
Conference dates |
July 5-7, 2023 |
Closed |
The meal package fee includes all lunches, dinners, and access to the social event and dinner on 6th of July.
The meal package fee for domestic students includes all coffee breaks.
Registrations will be confirmed (by e-mail) upon reception of the meal package fee.
|
|
Early registration |
Late registration |
||
|
|
1,5 days |
3 days |
1,5 days |
3 days |
|
ML4NGP MC members and non-members |
75 EUR |
150 EUR |
100 EUR |
200 EUR |
|
Domestic students |
50 EUR |
– |
||
- Each meal package fee must be paid separately.
- Your name and institution must be identified clearly.
- In case of cancellation, the registration fee will not be refunded.
- Participants with an accepted abstract will be considered eligible for reimbursement on a best effort basis.
abstract information
Abstract submissions are no longer accepted.
The abstract should be of maximum 400 words and have a brief and concise summary of the background/motivation for the study, method and result(s), as well as conclusion/statement of significance. The abstract is submitted through the submission form. Submitters may choose to have their submission consider for talk and poster or poster only. Those abstracts which are not selected for talk will be considered for a poster.
*A few poster abstracts will be selected to give a flash presentation during the symposium.
Presentation time
Short talks – Speaking time is 15 minutes plus 5 minutes discussion.
Invited talks – Speaking time is 25 minutes plus 5 minutes discussion.
Flash posters – Speaking time is 5 minutes.
Posters should have up to 90 cm (width) x 120 cm (height). Poster numbers will be communicated to authors in due course.
Poster presenters are requested to set up their posters in the designated area before the conference starts. Presenters are expected to be available near their posters during the assigned session.
Speakers are required to have their presentation ready on a USB key.
Your slides should be either in .pdf, .ppt or .pptx format.
Location & venue
The 1st ML4NGP Meeting 2023 will take a place at the Clarion Congress Hotel Bratislava.
BRATISLAVA
Bratislava is a small historical city, but largest in Slovakia and a youngest european metropolis with many shopping, dining and natural wonders. Find more about our beautiful city at the official webpage: https://bratislava.sk/.



VENUE
In Clarion Congress Hotel refined taste meets high functionality, modern interior with elegant use of space. Hotel offers unique, multifunctional conference rooms with modern technical equipment and variable configuration with elegant design for hosting international conferences.
It is easily accessible from Airport by public transport and within walking distance to downtown and all famous sights.
Zabotova 2
811 04 Bratislava, Slovakia
Tel. +421 2 5727 7000
ACCOMODATION
We would like to kindly suggest the Clarion Congress Hotel as the conference will be held in one of its conference halls.
PRE-event meeting
Tuesday, July 4
16:00 – 18:00
Registration | Welcome desk
18:00 – 20:00
ML4NGP Core Group meeting | Closed session
CONFERENCE PROGRAM
Invited Speakers

Dr. Andrey KAJAVA
Centre de Recherche en Biologie cellulaire, Montpellier, France

Prof. Miguel ANDRADE
Johannes-Gutenberg University of Mainz, Mainz, Germany

Dr. Sonia LONGHI
CNRS and Aix-Marseille University, Marseille, France

Prof. Salvador VENTURA
Universitat Autònoma de Barcelona, Barcelona, Spain

Prof. Peter TOMPA
VIB-VUB Center for Structural Biology, Brussels, Belgium
Wednesday, July 5
08:00 – 09:00
Registration | Welcome desk
SESSION I - NGP classification
9:00 – 09:10
Opening Session
Alexander Monzon
9:10 – 09:40
Targeting disordered dipeptide repeats in ALS/FTD: a new modality in drug development
Peter Tompa
» Short Talks
9:40 – 10:00
Function of low complexity regions in literature: using transformer-based model to encode information in word embeddings
Sylwia Szymanska
10:00 – 10:20
Small-angle scattering for nonglobular proteins : perspectives of machine learning
Sergei Grudinin
10:20 – 10:40
Identifying novel short linear motifs based on a generalized machine-learning based bioinformatic pipeline
Matyas Pajkos
10:40 – 11:10
Coffee Break
SESSION II - Intrinsic Disorder
11:10 – 11:40
Intrinsic disorder and amyloid-like fibril formation by the Henipavirus V and W proteins
Sonia Longhi
» Short Talks
11:40 – 12:00
Mechanisms underlying functional selection in intrinsically disordered proteins
Lucia Beatriz Chemes
12:00 – 12:20
Decoding the Black Box of protein language models: A Basic Analysis of Sequence Features and Protein Disorder Encoding
Zsuzsanna Dosztányi
12:20 – 12:40
Detailed insight into dynamics of intrinsically disordered proteins using high-resolution relaxometry
Pavel Kadeřávek
12:40 – 14:00
Lunch Break
SESSION III - Machine Learning
14:00 – 14:30
Predicting Protein Aggregation: From Traditional Methods to Machine Learning
Andrey Kajava
» Short Talks
14:30 – 14:50
Defining and predicting conformational variability of proteins
Wim Vranken
14:50 – 15:10
A unified computational method with reinforcement learning for modeling conformational ensembles of complex protein architectures
Juan Cortes
15:10 – 15:30
Will the widespread use of AI-based structure prediction impact conformational ensemble use and ensemble generation?
Tamas Lazar
15:30 – 16:00
Coffee Break
SESSION IV - Protein Repeats
16:00 – 16:30
Evolutionary study of protein Short Tandem Repeats in protein families
Miguel Andrade
» Short Talks
16:30 – 16:50
Resolving the Fine Structure in the Energy Landscapes of Repeat Proteins
R. Gonzalo Parra
16:50 – 17:10
Trimeric Autotransporters: non-globular, repetitive virulence factors with peculiar genomic recombination mechanisms
Dirk Linke
17:10 – 17:30
Evolutionary splicing graphs for matching non-globular protein sequences and describing protein repeats alternative usage in evolution
Elodie Laine
18:30 – 20:00
Poster session and welcome reception
Thursday, July 6
08:00 – 09:00
Registration | Welcome desk
SESSION V - NGP aggregation
09:00 – 09:30
Structural determinants driving oligomer to fibril conversion in an amyloid disease
Salvador Ventura
» Short Talks
09:30 – 09:50
dGAE(297-391) tau fragment promotes formation of CTE-like full-length tau filaments
Kristaps Jaudzems
09:50 – 10:10
AmyloGraph: A comprehensive database of amyloid-amyloid interactions
Michal Burdukiewicz
10:10 – 10:30
Cryo-EM structure of hnRNPDL-2 fibrils, a functional amyloid associated with limb-girdle muscular dystrophy D3
Javier Garcia Pardo
10:30 – 11:00
Coffee Break
» Short Talks
11:00 – 11:20
Non-globular protein based biomaterials and therapeutics by design
Esra Yüca Yılmaz
11:20 – 11:40
Mapping the structural transitions of the flanking polyQ regions in huntingtin exon 1 upon membrane binding
Ana M. Melo
11:40 – 12:00
LEA4 proteins: How disordered are they?
Marija Vidovic
12:00 – 12:20
Energy Embedding Transformer Network: A Novel Approach to Predict Functional Disorder in Proteins
Gábor Erdős
12:20 – 14:00
Lunch Break
14:00 – 15:10
Flash Poster Presentations
» Short Talks
15:10 – 15:30
A systematic evaluation of AlphaFold-multimer for predicting LIR motif-Atg8 interactions
Vasilis Promponas
15:30 – 16:00
Coffee Break
16:00 – 17:15
WG1 meeting
hosted by Pavel Kadeřávek (WG1 leader)
17:15 – 17:45
WG5 meeting
hosted by Rita Vilaça (WG5 leader)
19:30 – 22:00
Social event and dinner at the “Hostinec U Deda” restaurant (website)
Friday, July 7
9:00 – 10:15
WG2 meeting
hosted by Zsuzsanna Dosztányi (WG2 leader)
10:15 – 10:45
Coffee Break
10:45 – 12:30
MC meeting
12:30 – 14:00
Lunch
14:00 – 15:15
WG3 meeting
hosted by Jovana Kovačević (WG3 leader)
15:15 – 16:30
WG4 meeting
hosted by R. Gonzalo Parra (WG4 leader)
ORGANIZATION
SCIENTIFIC COMMITTEE
Alexander Monzon | Italy
Zuzana Bednarikova | Slovakia
Silvio Tosatto | Italy
Rita Vilaça | Portugal
Darius Sulskis | Lithuania
Pavel Kaderavek | Czech Republic
Zsuzsanna Dosztanyi | Hungary
Jovana Kovacevic | Serbia
Gonzalo Parra | Spain
LOCAL ORGANIZING COMMITTEE
Institute of Neuroimmunology SAS
Ondrej CEHLÁR
Rostislav ŠKRABANA
Gabriela KALAFUSOVÁ
Stefana NJEMOGA
Klaudia MEŠKOVÁ
Marián HORVÁTH
contact information
Information and inquiry: info@ml4ngp.eu
CONSORTIA

sponsor

This event is part of the activities of the COST Action ML4NGP, CA21160, supported by COST (European Cooperation in Science and Technology).
FEEDBACK FROM PARTICIPANTS
The presentations were very interesting and very helpful for my research in the future.
Great program/speakers, adequate time for discussions/networking.
Good balance between experimental/computational talks and the scientific networking.
I really enjoy the breaks, poster session and dinner because that's when you get to know your colleagues and discuss about their current work and future perspectives.
